Phylogenetic trees can be imported into PASSaGE from the Newick format. The data files are simply text files, but unlike the standard text file import, the data is not stored in rows and columns. The Newick format is a commonly used means of describing trees using parentheses and commas. For example:
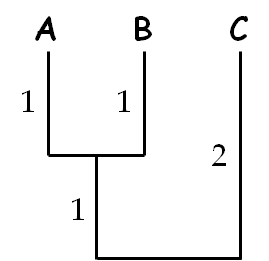
((A:1,B:1):1,C:2):0;
describes a three-taxon tree.

The phylogenetic tree described by “((A:1,B:1):1,C:2):0;”
Numbers which appear after a colon (“:”) indicate branch lengths. The end of the tree is indicated by a semi-colon (“;”), allowing large trees to be split over multiple lines. Taxon names can be of any length and can contain any character other than commas, colons, semi-colons, and open and close parentheses, since these symbols are used to describe tree shape. By default, any unspecified branch length will be assumed to have a length of zero.
If the terminals of a phylogeny are to be associated with a data matrix, the order is very important. Taxon order is described as found in the tree and will be used to match to a matrix. Thus, while
((A,B),C); and (C,(A,B));
describe the identical tree, PASSaGE will treat the taxon order in the first case as taxon 1 = A, taxon 2 = B, and taxon 3 = C, while in the second case it would find taxon 1 = C, taxon 2 = A, and taxon 3 = B.
PASSaGE can also read trees stored in Nexus format by programs such as PAUP*. Only the first tree will be loaded if more than one tree is stored in the trees block of the Nexus file. Trees in Nexus format may have a translation table where each taxon name is associated with a specific number and the numbers are used within the parenthetical format rather than the names directly.